(Comments)

In this tutorial, we are going to build a model to triage based on patient queries text data.
For example
| query (input) | triage (output) |
| Skin is quite itchy. | dermatology |
| Sore throat fever fatigue. | mouthface |
| Lower back hurt, so painful. | back |
We are going to use Keras with Tensorflow (version 1.3.0) backend to build the model
For source code and dataset used in this tutorial, check out my GitHub repo.
Python 3.5, numpy, pickle, keras, tensorflow, nltk, pandas
1261 patient queries, phrases_embed.csv came from Babylon blog "How the chatbot understands sentences".
Check out the data visualization here.
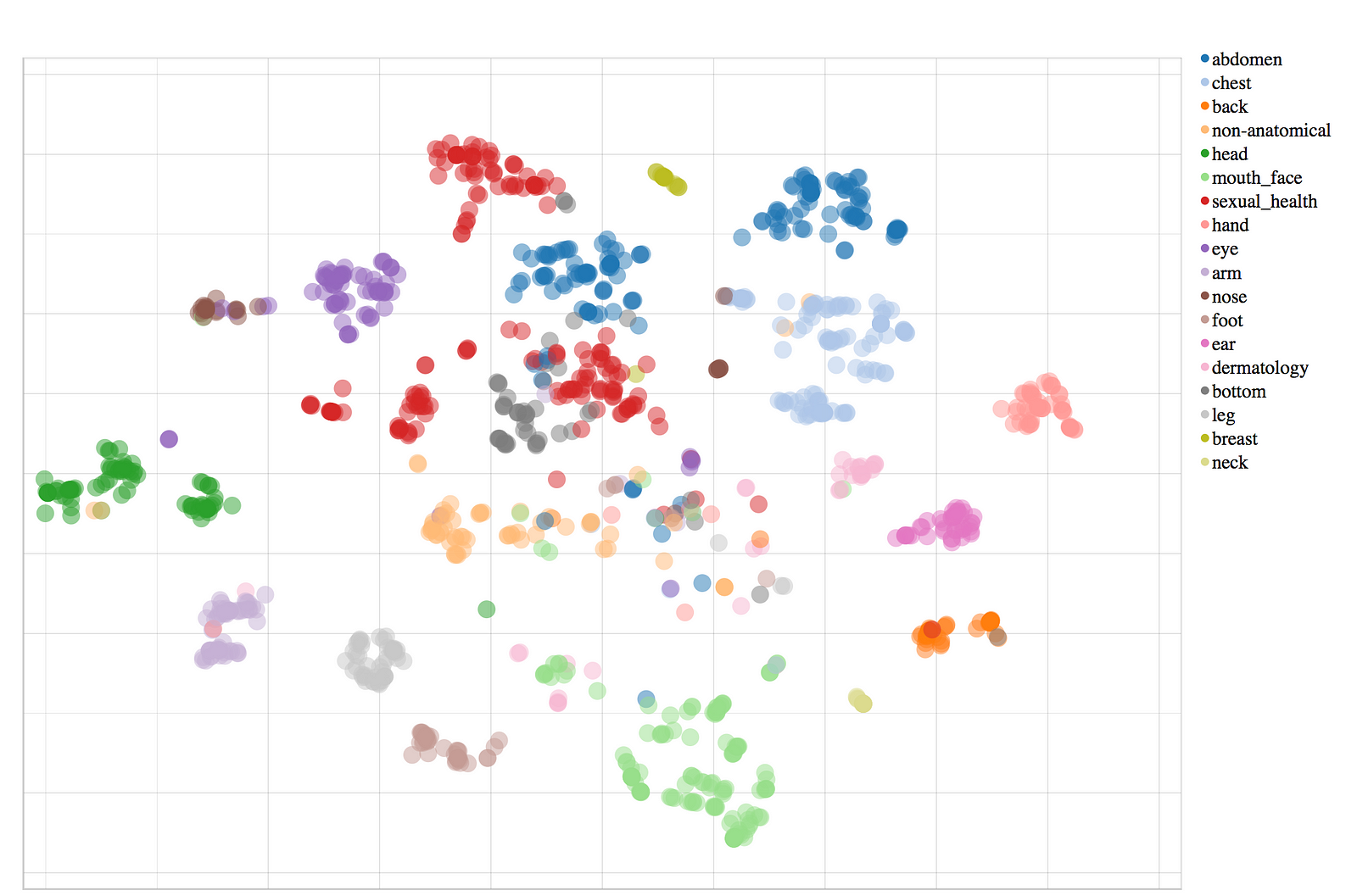
We will be doing the following steps to prepare data for training the model.
1. Read the data from CSV file to Pandas data frame, only keep 2 columns "Disease" and "class"

2. Convert Pandas data frame to numpy arrays pairs
"Disease" columns ==> documents
"class" columns ==> body_positions
3. Clean up the data
For each sentence, we convert all letter to lower case, only keep English letters and numbers, remove stopwords as shown below.
strip_special_chars = re.compile("[^A-Za-z0-9 ]+")
def cleanUpSentence(r, stop_words = None):
r = r.lower().replace("<br />", " ")
r = re.sub(strip_special_chars, "", r.lower())
if stop_words is not None:
words = word_tokenize(r)
filtered_sentence = []
for w in words:
if w not in stop_words:
filtered_sentence.append(w)
return " ".join(filtered_sentence)
else:
return r
4. Input tokenizer converts input words to ids, pad each input sequence to max input length if it is shorter.
Save the input tokenizer since we need to use the same one to tokenize any new input data during prediction.
5. Convert output words to ids then to categories(one-hot vectors)
7. Make target_reverse_word_index to turn the predicated class ids to text.
The model structure will look like this
_________________________________________________________________ Layer (type) Output Shape Param # ================================================================= embedding_1 (Embedding) (None, 18, 256) 232960 _________________________________________________________________ gru_1 (GRU) (None, 18, 256) 393984 _________________________________________________________________ gru_2 (GRU) (None, 256) 393984 _________________________________________________________________ dense_1 (Dense) (None, 19) 4883 =================================================================
The embedding layer transforms words ids into their corresponding word embeddings, each output from embedding layer would have a size of ( 18 x 256) which is the maximum input sequence padding length times embedding dimension.
The data is pass to a recurrent layer to process the input sequence, we are using GRU here, you can also try LSTM.
All the intermediate outputs are collected and then passed on to the second GRU layer.
The output is then sent to a fully connected layer that would give us our final prediction classes. We are using "softmax" activation to give us a probability for each class.
Use standard 'categorical_crossentropy' loss function for multiclass classification.
Use "adam" optimizer since it adapts the learning rate.
model = Sequential()
model.add(Embedding(vocab_size, embedding_dim,input_length = maxLength))
model.add(GRU(256, dropout=0.9, return_sequences=True))
model.add(GRU(256, dropout=0.9))
model.add(Dense(output_dimen, activation='softmax'))
tbCallBack = TensorBoard(log_dir='./Graph/medical_triage', histogram_freq=0,
write_graph=True, write_images=True)
model.compile(loss='categorical_crossentropy', optimizer='adam', metrics=['accuracy'])
We will then train the model and save it for later prediction.
1. Load the model we save earlier.
2. Load the input tokenizer and tokenize a new patient query text, pad the sequence to max length
3. Feed the sequence to model, the model will output the class id along with the probability, we use "target_reverse_word_index" to turn the class id to actual triage result text.
Here are some predicted result

Keras trained for 40 epochs, takes less than 1 minute with GPU (GTX 1070) final acc:0.9146
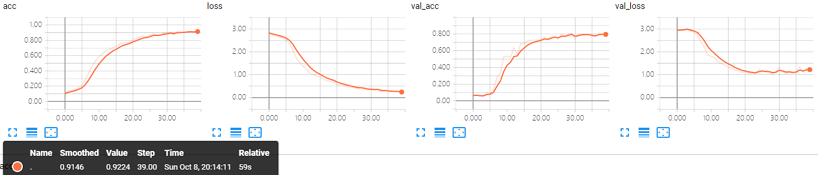
The training data size is relatively small, having larger datasets might increase the final accuracy.
Check out my GitHub repo for the Jupyter notebook source code and dataset.
Share on Twitter Share on Facebook
Comments